
RBCs are well suited for this immune defense role, having no nucleus or other infrastructure to perform viral replication and being expendable, numbering 20–30 trillion per human host.
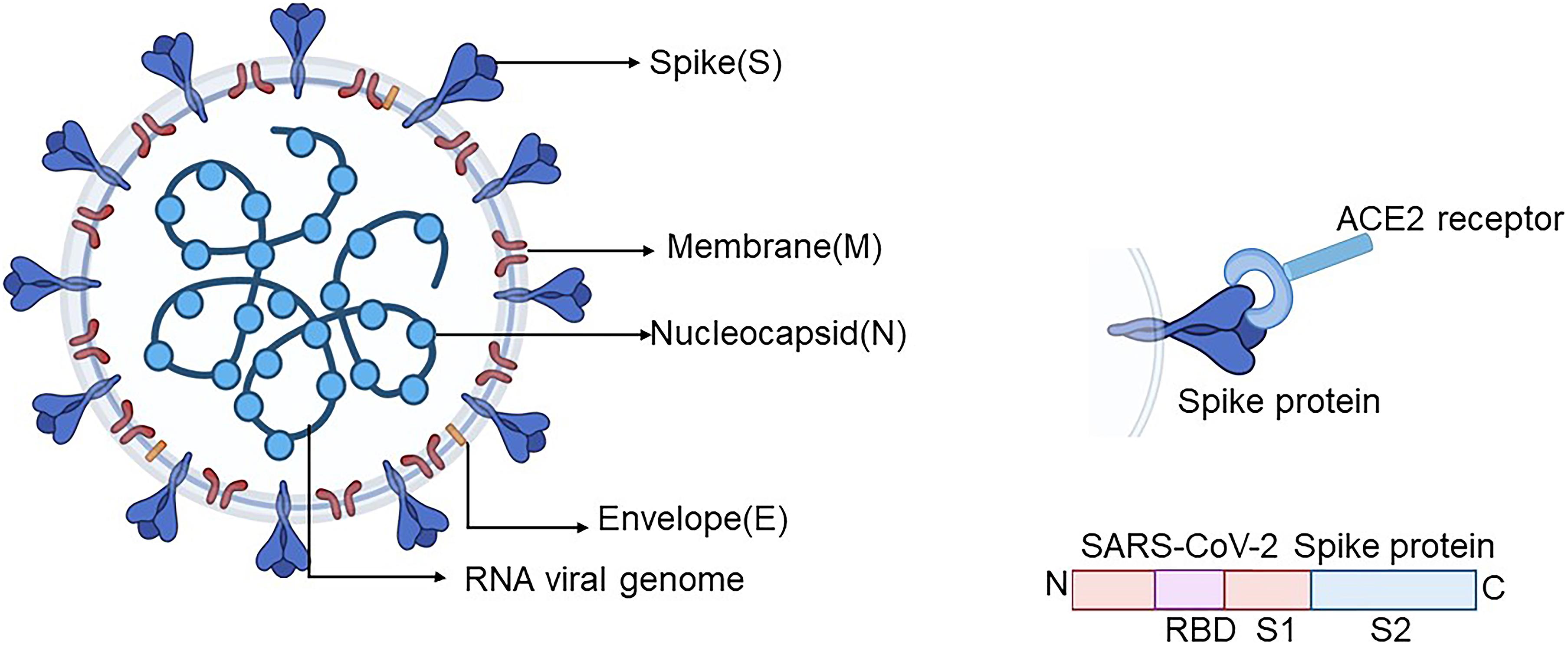
Viruses, however, are snagged directly by RBCs, and viral–RBC binding is inhibited by antiviral antibodies. With bacteria, this clearance by RBCs requires both antibody and complement. RBCs perform an active pathogen clearance role, attaching to microorganisms and then delivering them to leukocytes or conveying them to macrophages in the liver or spleen for phagocytosis. In particular, SA moieties are densely distributed on the RBC surface (35 million per cell ), mainly as terminal residues of both the CD147 receptor and GPA, with GPA serving no known physiological role other than as a decoy for pathogens. The human host, reciprocally, protects against attachment of virions to host cell infectious targets by presenting SA on a set of decoys, including RBCs as well as platelets and leukocytes. Binding of Viruses to SA and Host Decoy Defenseįor viruses that bind to SA, including SARS-CoV-2 as noted above, such glycan bindings play a key role in viral infectivity, as SA typically functions as an initial point of attachment to host cells. Figure 1 shows a schematic of virally mediated hemagglutination and of its inhibition through an agent that competitively binds to attachment sites on the virus, in this case an antiviral antibody.ġ.1. However, for viruses that express enzymes that cleave SA, that sheet subsequently collapses as these enzymes disintegrate SA-binding sites on the RBCs. In that assay, developed in the 1940s and refined later by Jonas Salk, virus particles are mixed with RBCs to form a hemagglutinated sheet. Hemagglutination also occurs for several other viral strains including other coronaviruses, as demonstrated in the classic viral hemagglutination assay. Through viral bindings to SA surface moieties to be considered in detail below, SARS-CoV-2 agglutinates RBCs, as established in vitro, with such bindings also demonstrated clinically. SA is densely distributed on red blood cells (RBCs) as terminal residues of its surface sialoglycoprotein, glycophorin A (GPA), and of its CD147 transmembrane receptors. However, many enveloped viral strains, including coronaviruses, initially attach to host cell membranes via glycoconjugate molecules, including those tipped with sialic acid (SA).
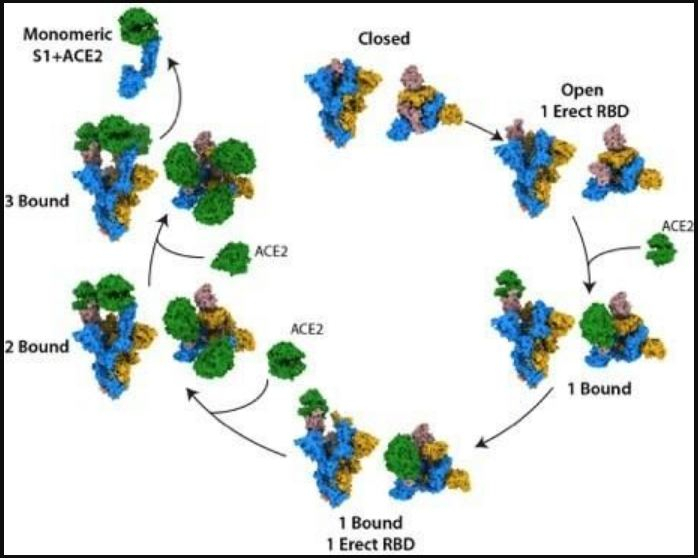
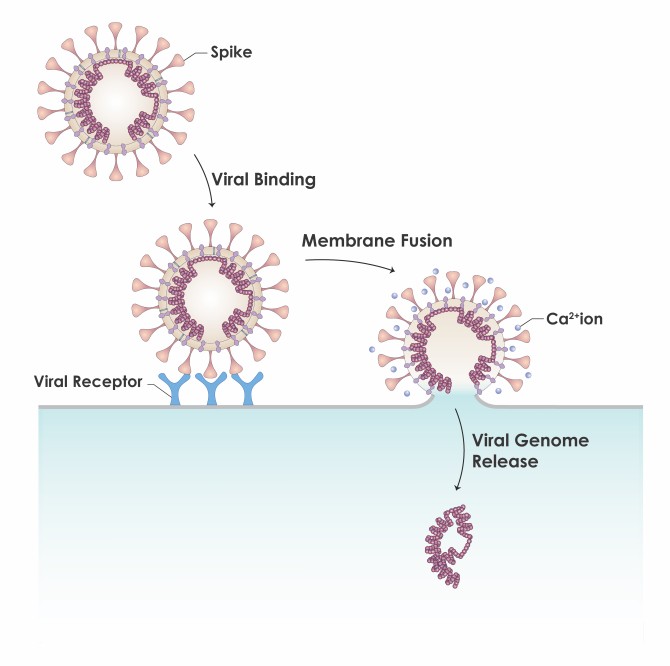
Viruses fuse and then replicate via host cell receptors specific to the viral strain, which is ACE2 for SARS-CoV-2. Ī framework for studying the vascular occlusive morbidities characteristic of COVID-19 is provided by viral properties dating back to classic experiments of the 1940s, as reviewed. One clinical reviewer characterized COVID-19 as “a systemic disease that primarily injures the vascular endothelium”. Histological studies from COVID-19 patients have found extensively damaged endothelium of pulmonary capillaries adjoining relatively intact alveoli, corresponding to hypoxemia with normal breathing mechanics observed in patients with this viral disease. The in vitro and clinical testing of these possibilities can be sharpened by the incorporation of an existing anti-COVID-19 therapeutic that has been found in silico to competitively bind to multiple glycans on SARS-CoV-2 spike protein.Īlthough COVID-19 typically gains infectious penetration in the respiratory epithelium, vascular damage is frequently observed in lungs and other organ systems of COVID-19 patients, with morbidities such as intravascular clotting, microvascular occlusion and peripheral ischemia. The arrangement and chemical composition of the glycans at the 22 N-glycosylation sites of SARS-CoV-2 spike protein and those at the sialoglycoprotein coating of RBCs allow exploration of specifics as to how virally induced RBC clumping may form. For COVID-19, key such properties are: (1) SARS-CoV-2 binds to RBCs in vitro and also in the blood of COVID-19 patients (2) although ACE2 is its target for viral fusion and replication, SARS-CoV-2 initially attaches to sialic acid (SA) terminal moieties on host cell membranes via glycans on its spike protein (3) certain enveloped viruses express hemagglutinin esterase (HE), an enzyme that releases these glycan-mediated bindings to host cells, which is expressed among betacoronaviruses in the common cold strains but not the virulent strains, SARS-CoV, SARS-CoV-2 and MERS. Rouleaux (stacked clumps) of red blood cells (RBCs) observed in the blood of COVID-19 patients in three studies call attention to the properties of several enveloped virus strains dating back to seminal findings of the 1940s.


 0 kommentar(er)
0 kommentar(er)
